Customizing interactive planning
Now, let’s try customizing the Interactive Path Planner (IPP) with a more complex molecule with a SMILES string of COc1ccc(C#CC2(O)C(C)CCCC2(C)C)cc1. In Modules>Retrosynthesis>Interactive Path Planning/Tree Builder, type in COc1ccc(C#CC2(O)C(C)CCCC2(C)C)cc1 as the Target.
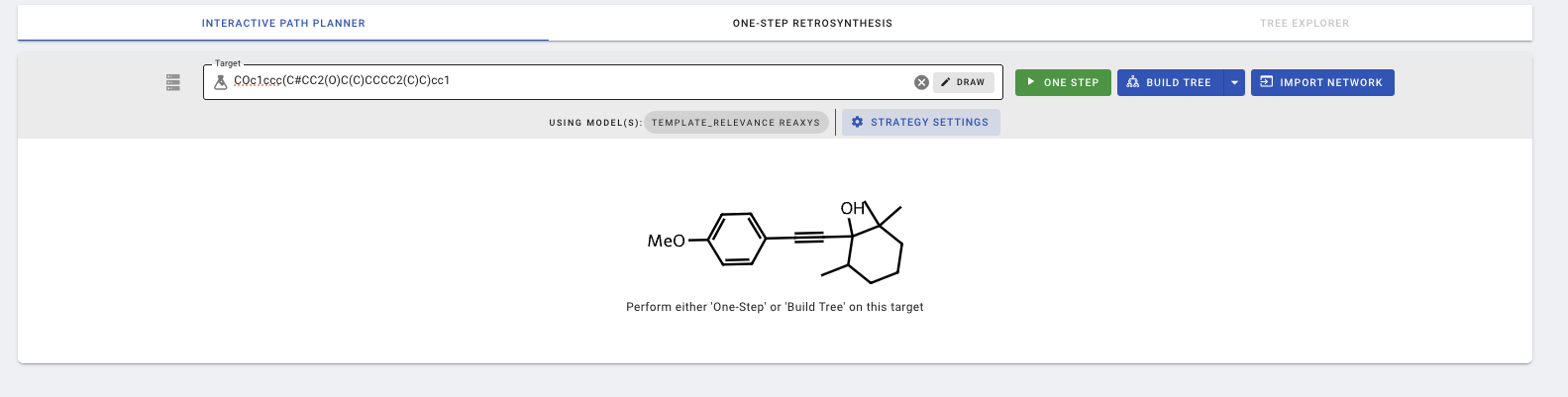
Before running the one step prediction, there are various settings in the IPP that the user can change, which can be accessed using the STRATEGY SETTINGS button. Most notably:
- Min. plausibility: Reaction plausibility is scored by the fast filter model, a data-driven model that scores a proposed reaction without considering reaction conditions. To maximize the number of returned suggestions and include less plausible predictions, you can set this value to 0.
- Apply regio-selectivity checking: When this setting is turned on, reactions are checked for regioselectivity. Reactions with two or more regio-selective products are flagged with red circles. Reactions where the model is unable to check regio-selectivity are flagged with yellow circles.
- Strategy Plan: The default strategy uses template_relevance model, which is a template-based one step model. This can be changed to augmented_transformer, which is a template-free model.
- template_relevance: Within the template_relevance model, we can change the template sets, maximum number of templates, and maximum cumulative probability. We can also combine several template sets by adding a Strategy Plan for the second (or more) template sets.
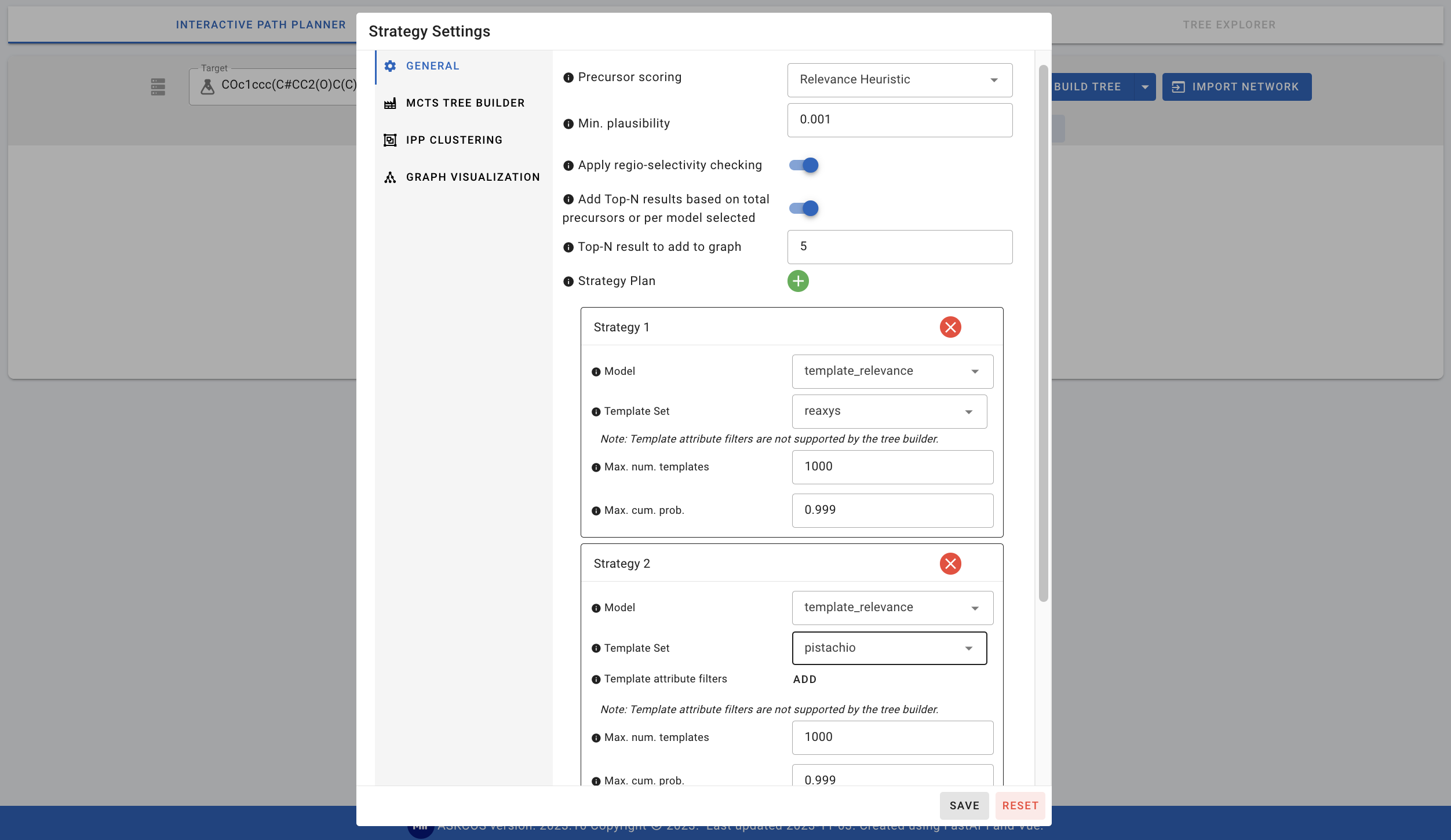
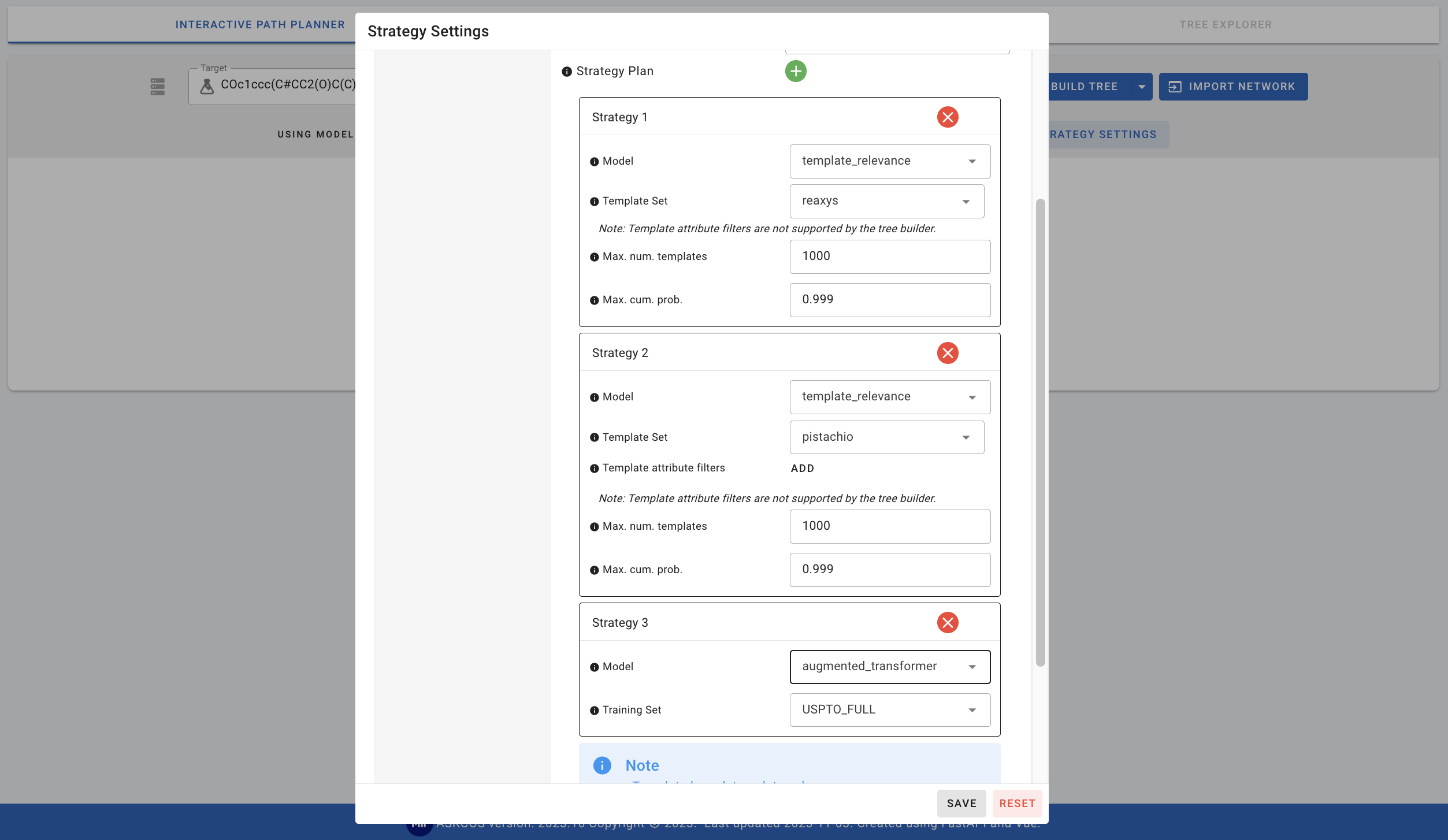
- augmented_transformer: The default training set is USPTO_FULL and this can be changed to pistachio_23Q3.
Note that the template_relevance and augmented_transformers can also be combined with each other. Let’s try combining template_relevance model with reaxys and pistachio template sets as well as the augmented_transformer with USPTO_FULL dataset. Let's also apply regio-selectivity checking.
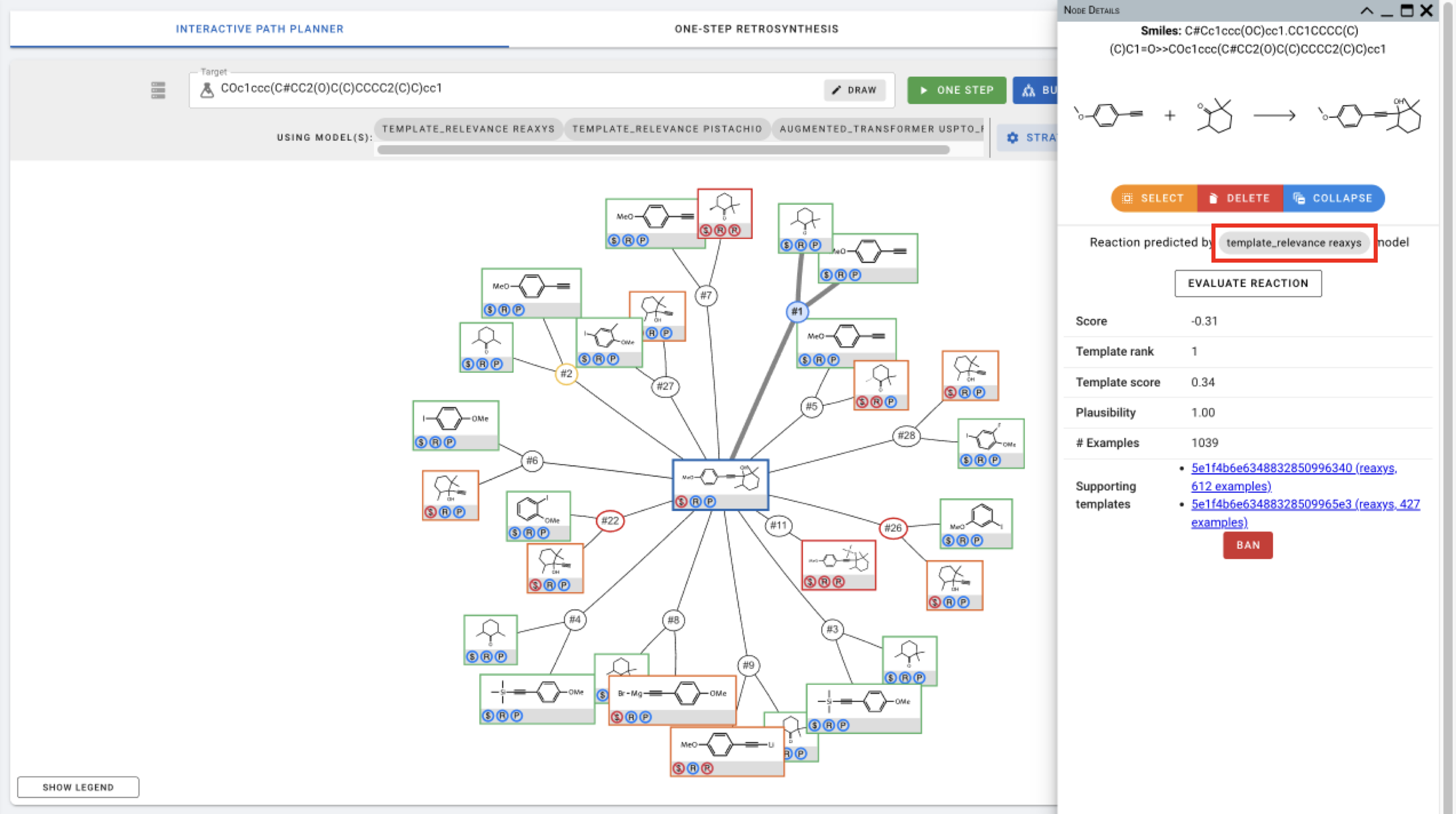
After adding these strategies, click SAVE and run ONE STEP. Running the one step prediction will give you 5 precursors (default setting) for each of the strategies. Once the predictions are complete, you can click on the individual reaction nodes to see which model the reaction is predicted by.
Understanding Node Details
- Chemical Nodes For unexpanded nodes, Node Details page will look like the following.
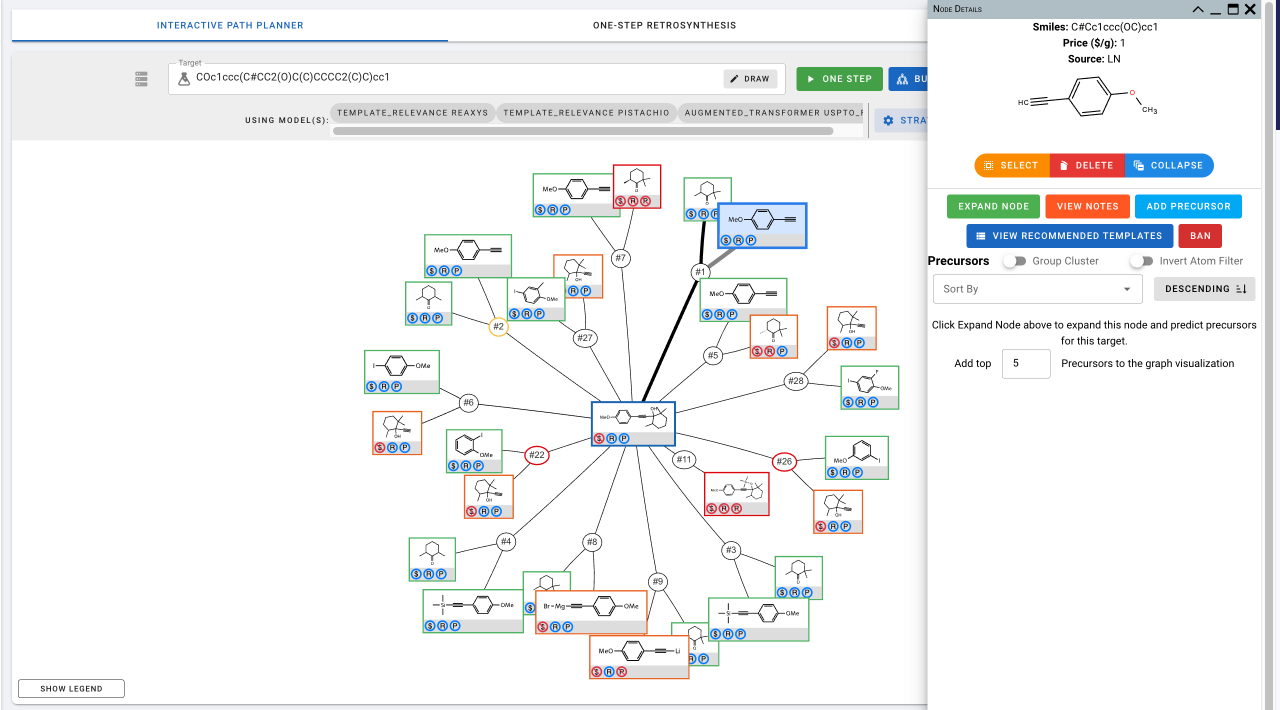
- Expand Node: You can run one step predictions for the selected molecule.
- View Notes: You can add and view notes about this molecule.
- Add Precursor: You can add any precursor(s) to this molecule. Once you have expanded a molecule, if the precursor(s) you are interested in is not included in the predictions, you can add them manually.
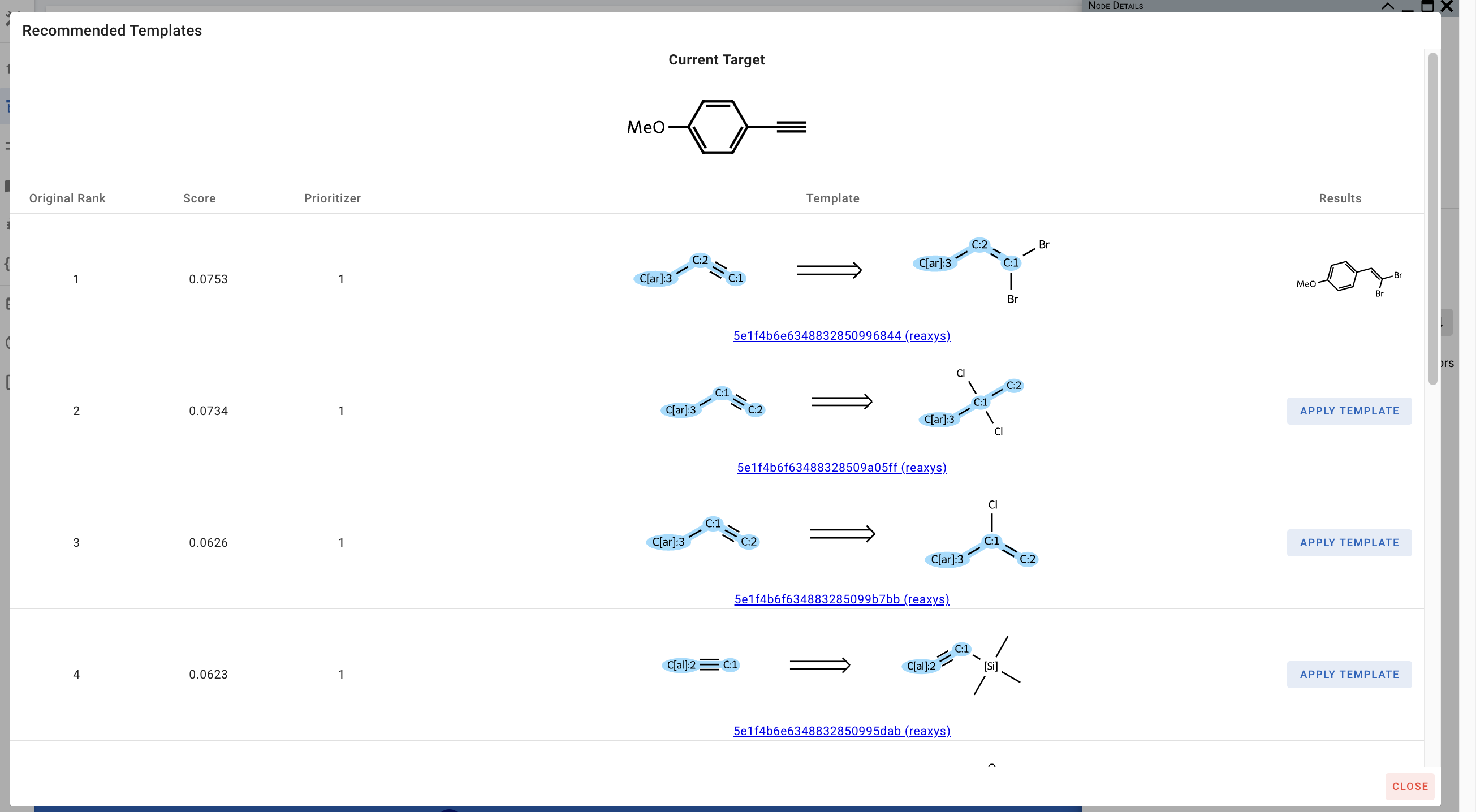
- View Recommended Templates: This shows you the templates that are recommended for this molecule and their associated scores/ranks. You can click on the link to view information about individual templates and the reactions they were extracted from. You can also view the results of applying each template to the current target.
- Ban: You can ban the specific chemical.
For expanded nodes including the target molecule, Node Details also include information about the predicted precursors for these nodes. You can sort the precursors using different criteria.
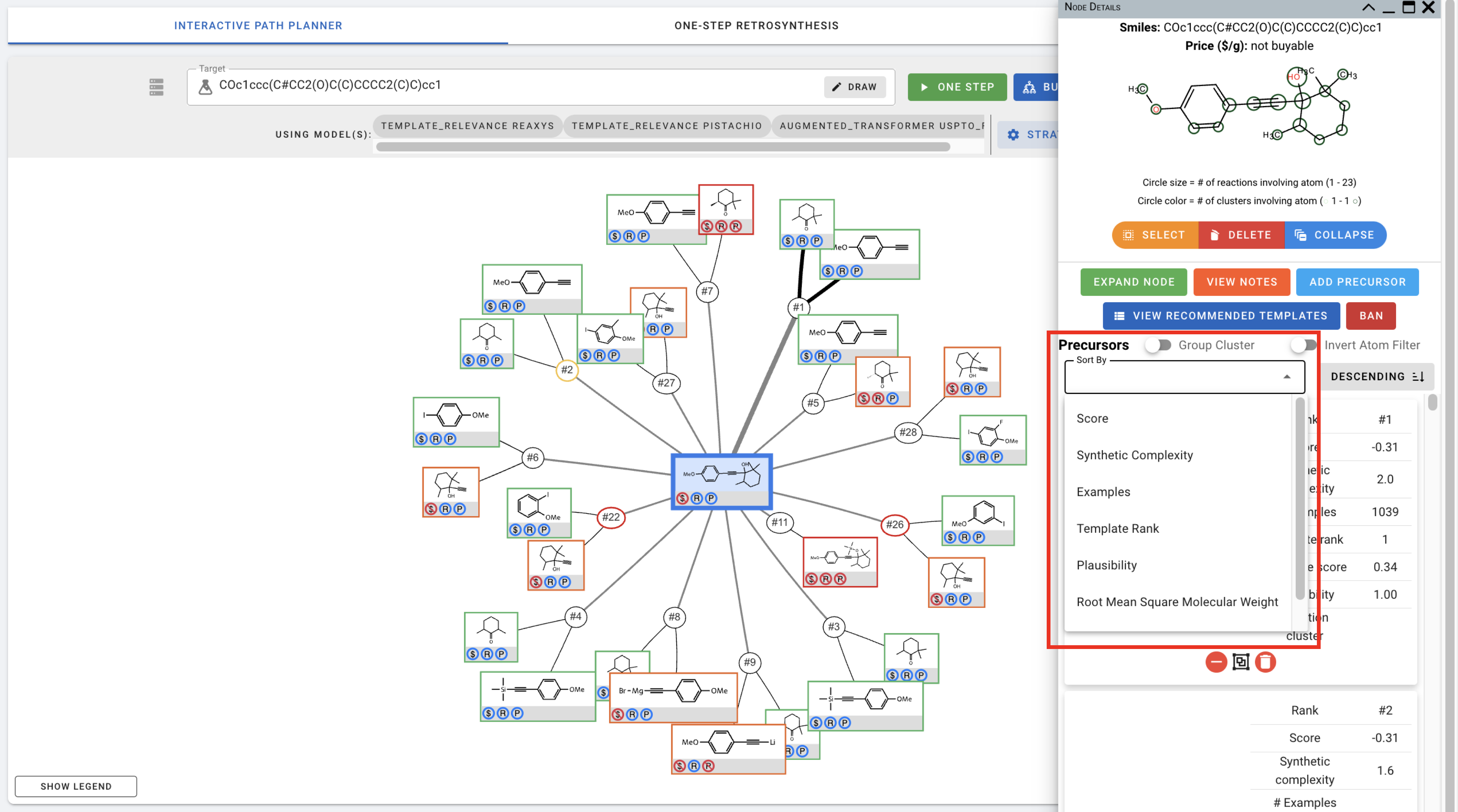
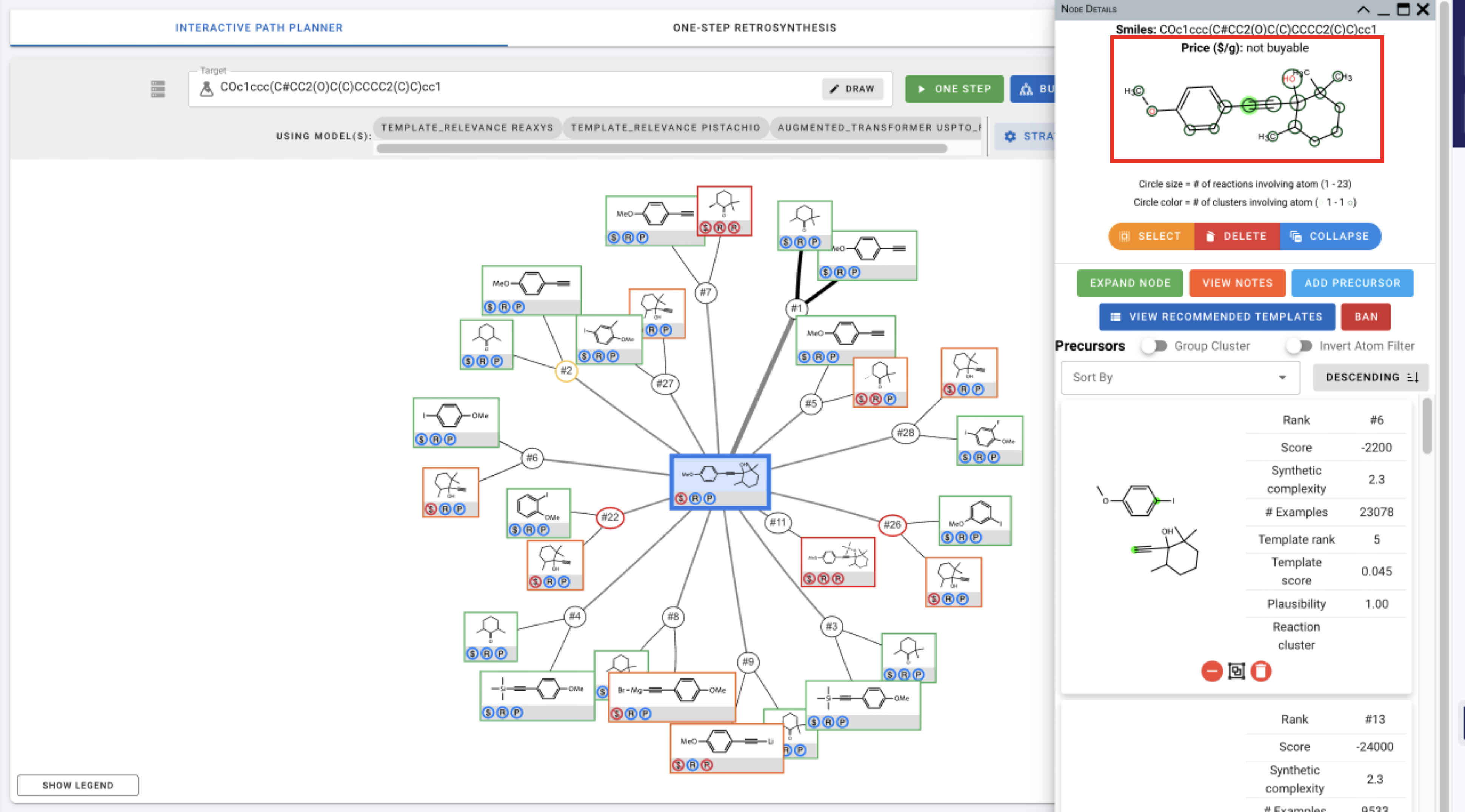
You can also click on different atoms in the figure on the top to view precursors that involve changes in the selected atom.
- Reaction Nodes
- template_relevance: Reaction nodes for reactions predicted by the template_relevance model gives you the score, template rank, template. score, plausibility, # of examples associated with the template, and the supporting templates (with links to template information).
- augmented_transformer: Reaction nodes for reactions predicted by the augmented_transformer gives you the score and plausibility of the reaction.
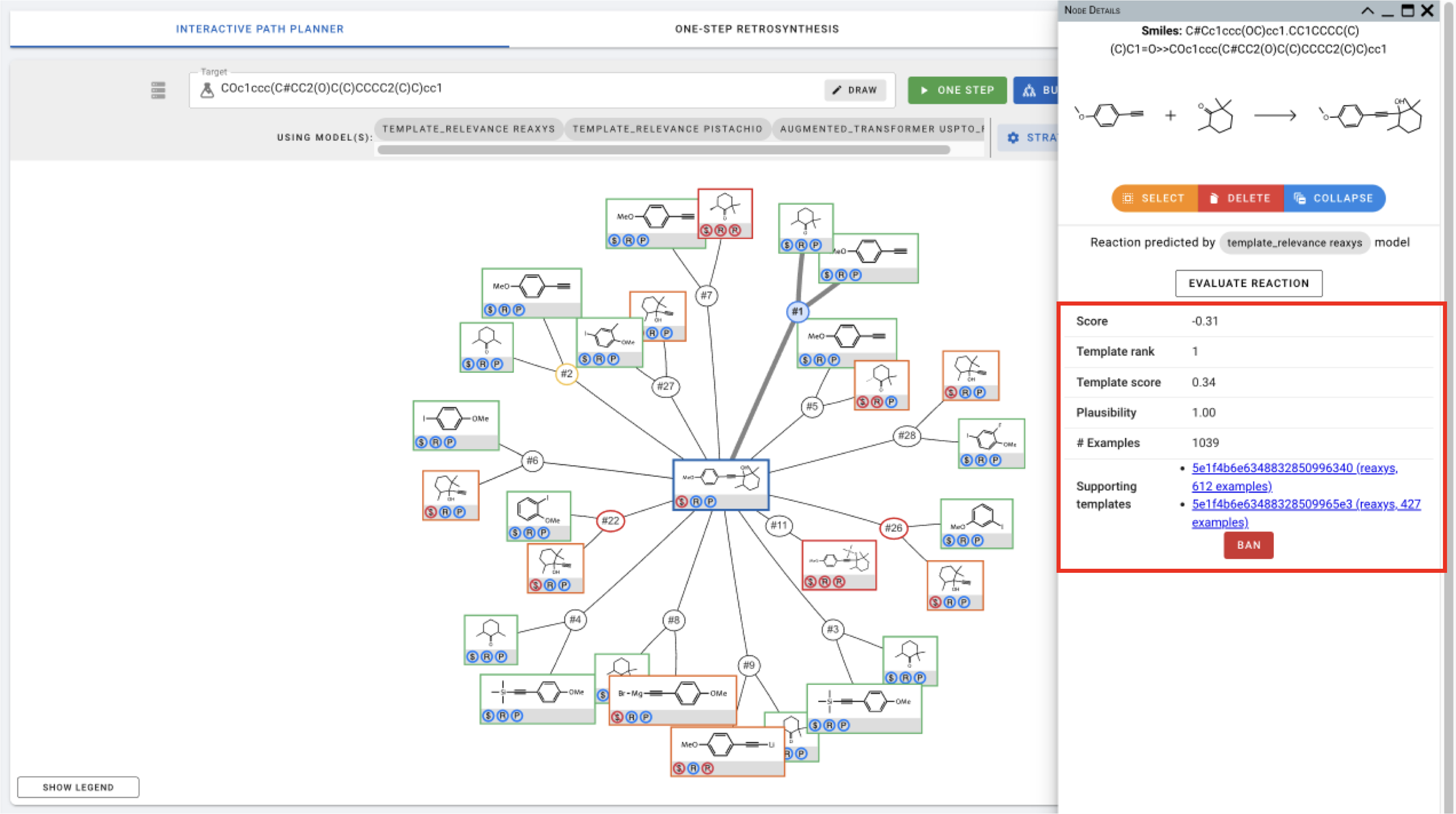
Click EVALUATE REACTION to move to forward synthesis analysis.
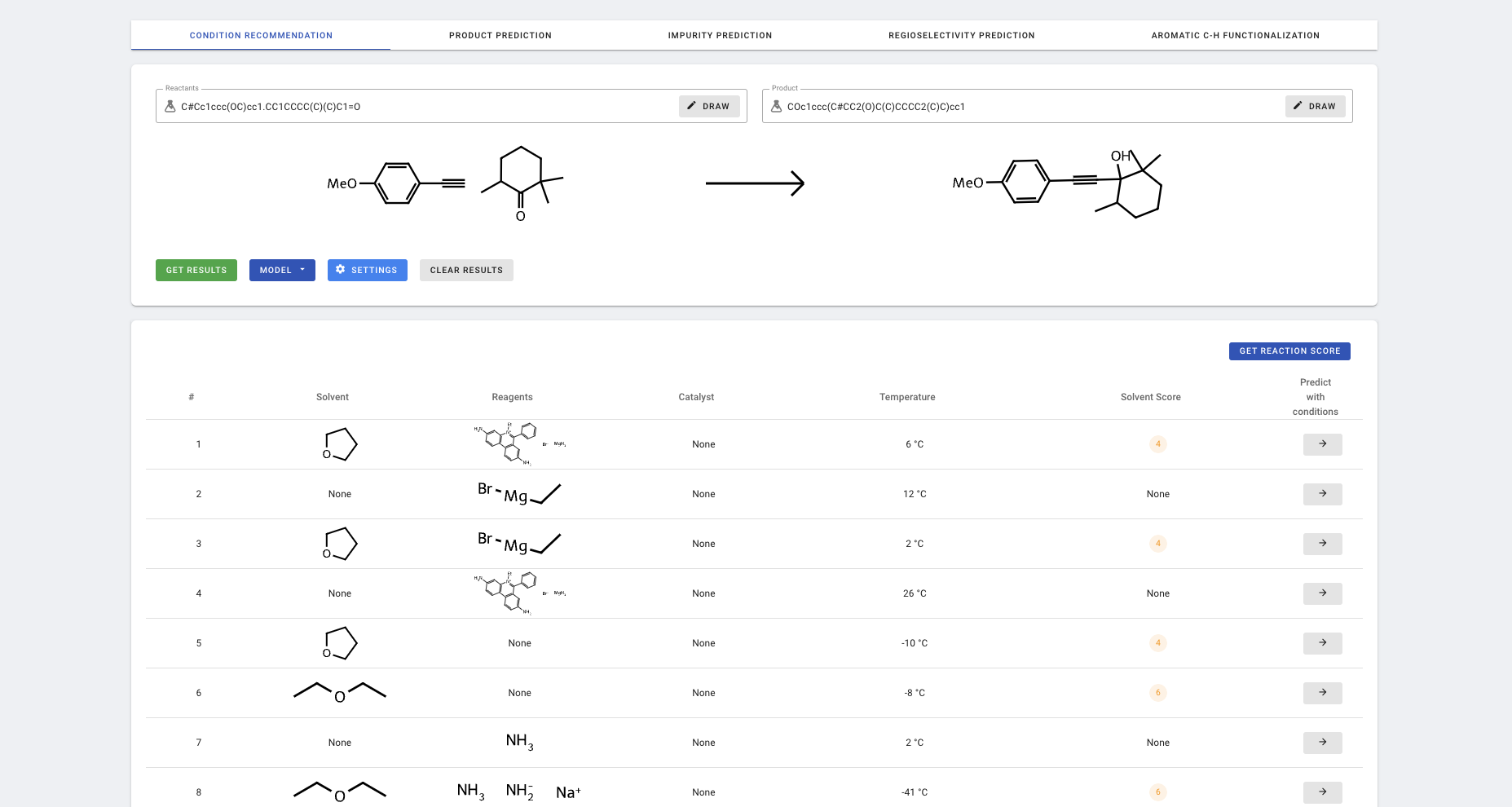
More information about forward synthesis analysis can be found in [05-Forward synthesis analysis](05-Forward synthesis analysis).