More tree analysis examples
In this section we will dive deeper into the analyses available for results returned by the Tree Builder.
We will continue working off of the results returned in the example from the previous section for the SMILES string: Cc1cc(O)c2c(c1)C(=O)c1cc(O)cc(O)c1C2=O.
Returned pathways can be viewed by clicking into VIEW TREES under the entry for the target molecule in MY RESULTS. This takes us to TREE EXPLORER mode.
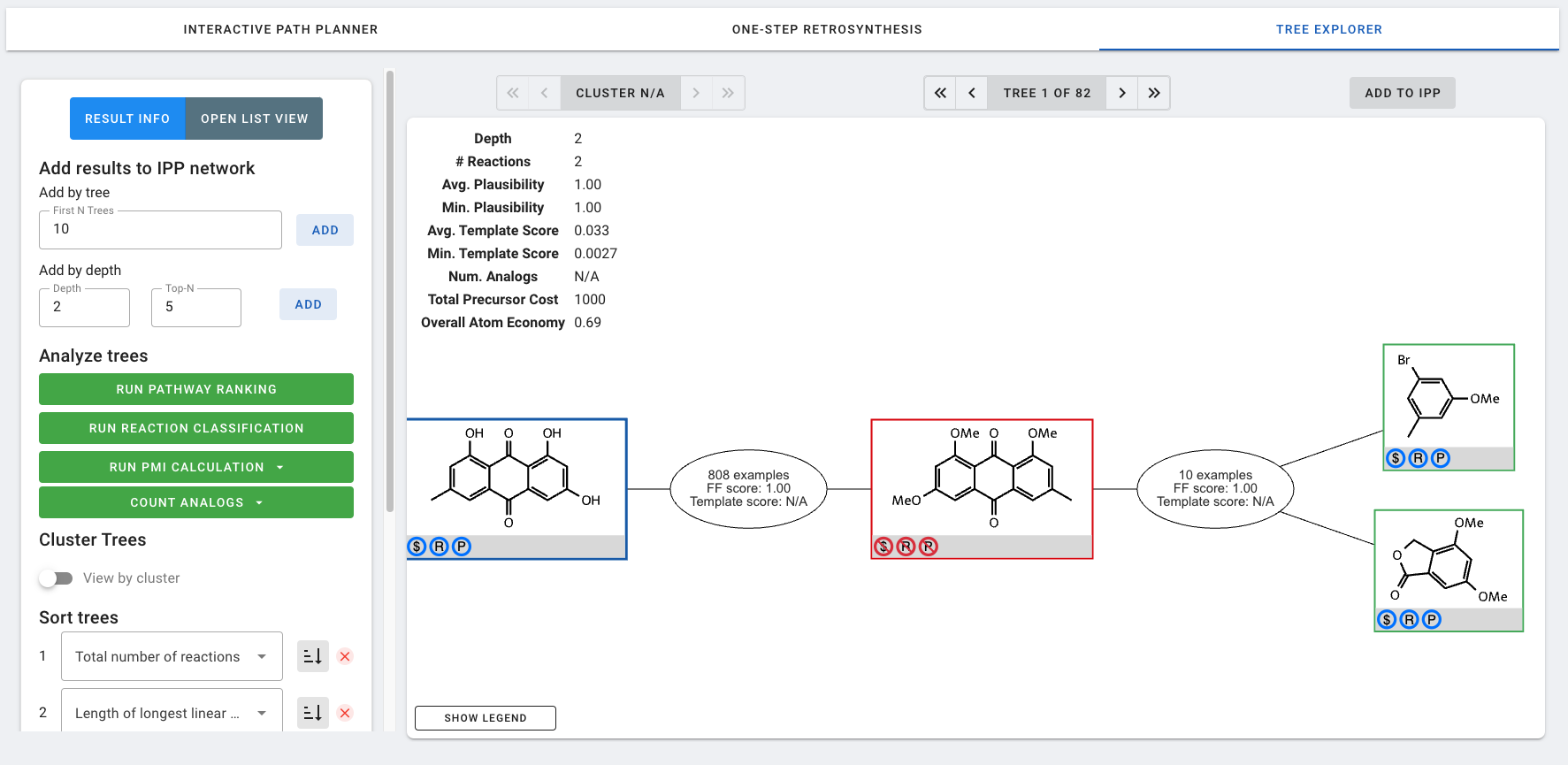
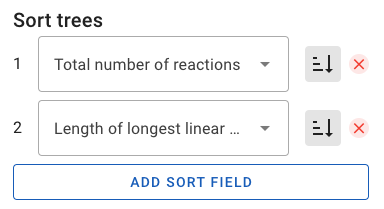
In TREE EXPLORER mode, you can navigate between the returned synthetic routes ("trees") using the arrows above the main display window. By default, trees are sorted from fewest to most reactions. This setting can be changed in the Sort trees settings. Whether the sorting is performed in ascending or descending order can be toggled by clicking on the icon to the right of the dropdown menu. Finally, additional secondary, tertiary, etc. sort fields can be added or removed
For each tree, information is displayed in the top left of the main display window. Additional parameters can be computed using the buttons in the Analyze trees section of the settings. These analyses will run in the background and will be displayed once they are completed and the window is refreshed.
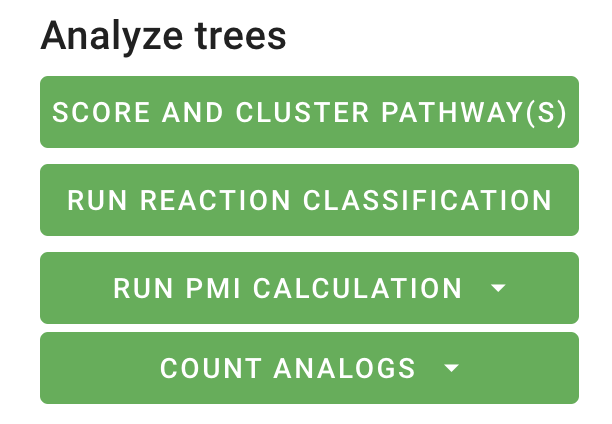
Score and cluster pathway(s): This analysis scores and clusters the pathways. Once this job is run, to increase the interpretability of the returned pathways--particularly when there is a large number of returned pathways--trees can be clustered and viewed by cluster by toggling the View by cluster setting.

This enables navigation to be performed between clusters and between trees in one cluster with the respective arrows above the main display window.

Run reaction classification: This analysis assigns a reaction class to each reaction in the pathway(s). Once this job is run, the reaction class details will be added to the node details for the reaction nodes in the pathway(s).
Run PMI calculation: This analysis calculates the process mass intensity (PMI) of the pathway(s). Once this job is run, the average PMI will be displayed in the top left corner of the main display window.
Count analogs: This analysis estimates the number of chemical analogs to the original product molecule are synthetically accessible given a synthetic route and the ASKCOS buyables compound database. Once this job is run, the number of analogs will be displayed in the top left corner of the main display window.
PMI calculation and count analogs can be run for all trees at once or for each tree individually. Running these analyses will also enable new sorting options in the sorting dropdown menu.