More forward synthesis analysis examples.
Five different machine learning models for forward synthesis are accessible from the side bar under Forward Synthesis. These are:
Condition Recommendation
Condition recommendation predicts optimal reaction conditions for specific reactions. Reactants and expected products are required for this prediction.
- Enter CC(=O)O.CCCO as the Reactants and CCCOC(C)=O as the Products, which are the SMILES for the mixture of acetic acid and propan-1-ol, and propyl acetate.
- Click Settings to choose the model.
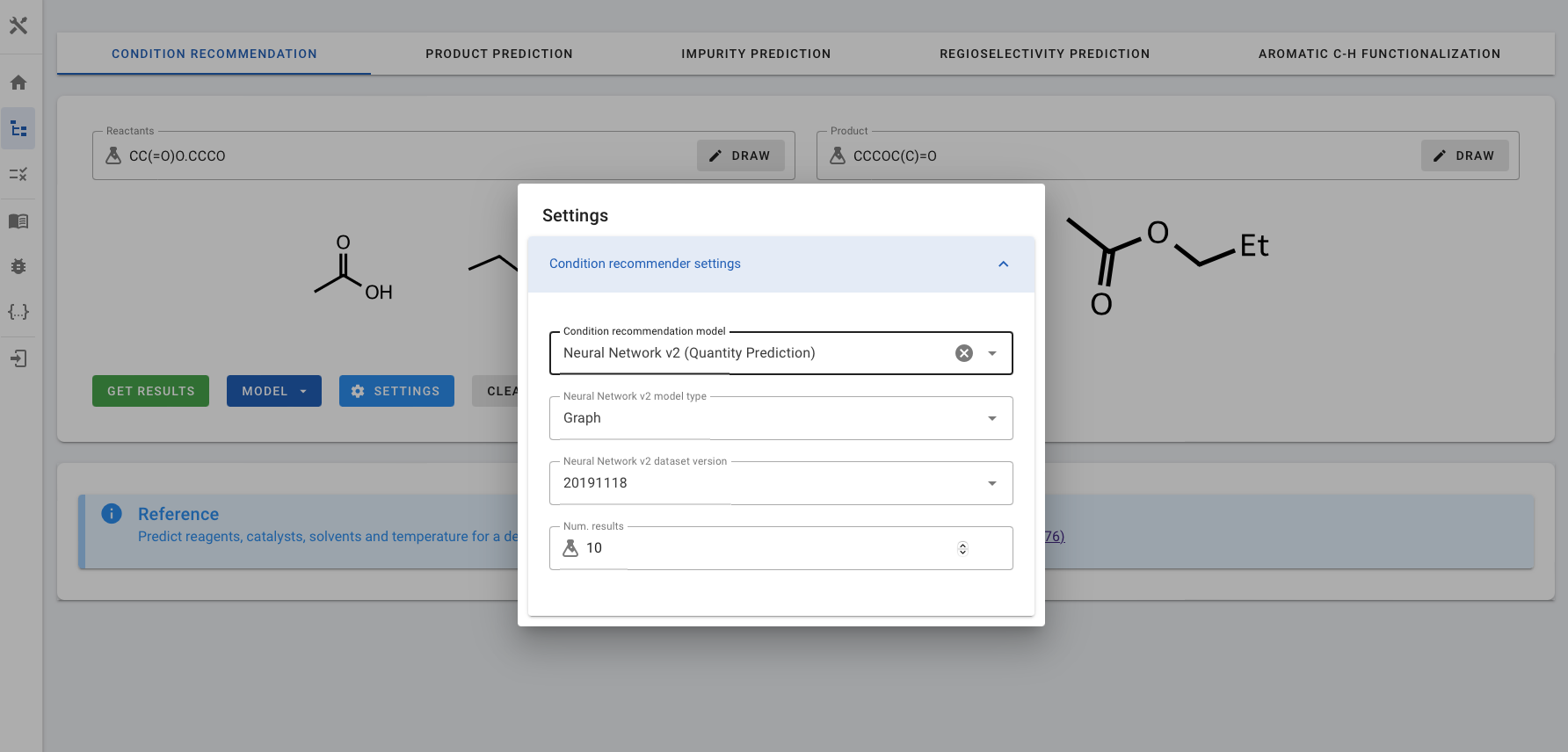
- Condition recommendation model: Choose the desired model for condition recommendation. There are two models available, the default is Neural Network.
- Neural Network: It predicts solvent, reagent, catalyst, temperature, and solvent score.
- Neural Network v2 (Quantity Prediction): It predicts the Reactants and Reagents with relative amounts (or stoichiometries), and temperature. You will then need to choose the model type and dataset version.
- Neural Network v2 model type: Select either the graph-based or fingerprint-based model, the default setting is Graph. NOTE - The graph model requires atom-mapped reactions. If atom-mapping is not available, please use the fingerprint model instead.
- Neural Network v2 dataset version: The only available dataset option is 'Pistachio 2019-11-18’, which is 20191118.
- Number of Recommendations: Adjust the number of recommendations to be displayed. The default setting is 10.
- Condition recommendation model: Choose the desired model for condition recommendation. There are two models available, the default is Neural Network.
- Click Get results to start the prediction. Get Reaction Score predicts the plausibilty of this reaction (Need to check).
In the example below, the default settings were used (Neural Network model). The results table lists the predicted reaction conditions based on rank.: 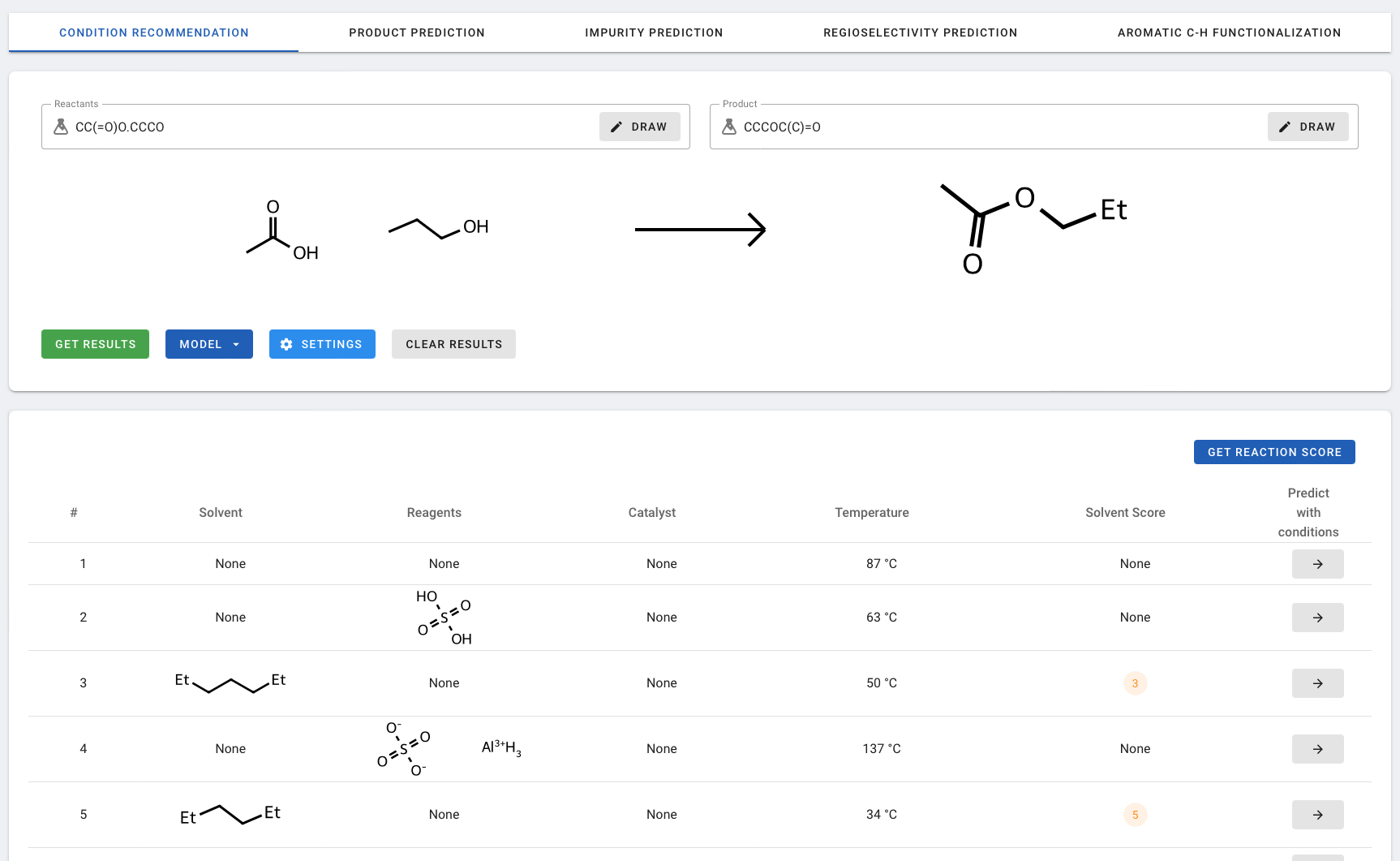
- #: The predicted ranking provided by the model
- Reagents, catalyst, solvents, and temperature: The reaction conditions predicted by the model.
- Solvent Score: Rates the Environment, Health, and Safety aspects of a solvent from 1 to 6, with 1 being most green and 6 being most hazardous. A score of 7 indicates no EHS information was available. NOTE - Solvent Score is only predicted by the Neural Network model, not by the Neural Network v2.
- Prediction with conditions: If you find any conditions that are suitable/interesting, you can proceed to predict reaction outcomes under the given conditions by clicking the arrow which redirects to the Product Prediction page.
Reference: ACS Cent. Sci., 2018, 4, 1465-1476
Product Prediction
Product Prediction predicts most likely outcomes of a chemical reaction from the reactants under given reaction conditions. Reactants are required to predict products, but reagents and solvents are optional.
- Enter CC(=O)O.CCCO as the Reactants which are the SMILES for the mixture of acetic acid and propan-1-ol.
- Click Settings to choose the model.
- Forward prediction model: Choose the desired model for prediction. The default setting is wldn5.
- wldn5: A template-free WLN model for predicting likely bond changes. Chem. Sci., 2019, 10, 370-377
- graph2smiles: A template-free Graph2SMILES model for end-to-end prediction. J. Chem. Inf. Model., 2022, 62 (15), 3503–3513
- augmented_transformer: A template-free transformer model for end-to-end prediction. ACS Cent. Sci., 2019, 5 (9), 1572–1583
- Forward model training set: Select the dataset used to train the model. The default is pistachio for wldn5, and uspto-stereo for graph2smiles and augmented_transformer.
- Forward prediction model: Choose the desired model for prediction. The default setting is wldn5.
- Click Get results to start prediction.
The results table lists the potential products based on rank: 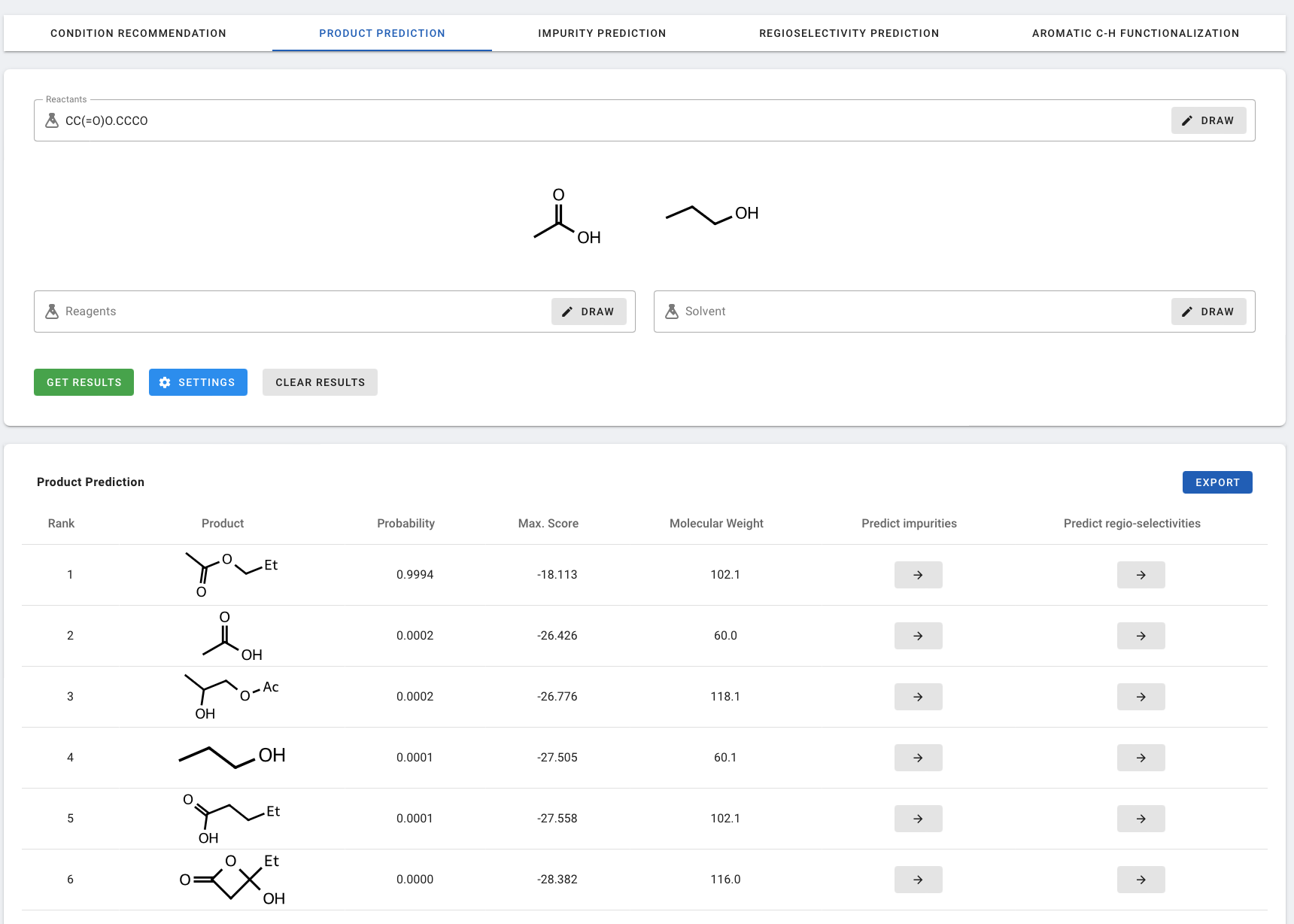
- Rank: The predicted ranking provided by the model.
- Product: The molecular structure of predicted product.
- Probability: The predicted probability of being a product for the given reactants.
- Max. Score: Reflects the fact that multiple graph edit sets could lead to the same (not-atom-mapped) product SMILES, and results are merged from the ranking model that have identical product SMILES as a post-processing step.
- Molecular Weigt: The molecular weight of the product.
- Predict impurities: Clicking the arrow redirects to Impurity Prediction page to predict the possible impurities of the reaction.
- Predict regio-selectivities: Clicking the arrow redirects to Regioselectivity Prediction page to predict the probabilities of the same type of reaction occurring at different reaction sites on the molecule.
Impurity Prediction
ASKCOS impurity predictor considers the following 5 modes of impurity formation.
- Minor product; inputs are reactants, outputs are Top-k predicted products of Product Prediction.
- Over-Reaction; inputs are reactants and products (could be one of the products of mode 1), outputs are overreaction impurities.
- Dimerization; inputs are two monomers (could be one of the products of mode 1 or reactants), outputs are dimer products.
- Solvent adduct; inputs are reactants or products (could be one of the products of mode 1) and solvent, outputs are solvent adduct products
- Subset of reactants; input is a subset of reactants, outputs are predicted products of Product Prediction.
To predict possible impurities,
- Enter CC(=O)O.CCCO as the Reactants and CCCOC(C)=O as the Products, which are the SMILES for the mixture of acetic acid and propan-1-ol, and propyl acetate.
- Click Settings to choose the model.
- Top-k from forward prediction: Number of predicted products from Mode 1 that will be considered in impurity prediction (Mode 2-4). Default is 3.
- Inspection threshold: Products/impurities from reactions with lower-than-threshold inspector scores will be filtered out. Default is 0.1.
- Inspector Score Selection: Choose the desired forward prediction model that will be used to predict outcomes of Mode 1-5. Defualt is Reaxys inspector.
- Use atom mapping: When turned on, Mode of impurity formation will be confirmed for each reaction by mapping the reaction and checking a reactant substructure is present in the product. Default is Off.
- Click Get results to start prediction.
The results table lists the predicted impurities based on rank: 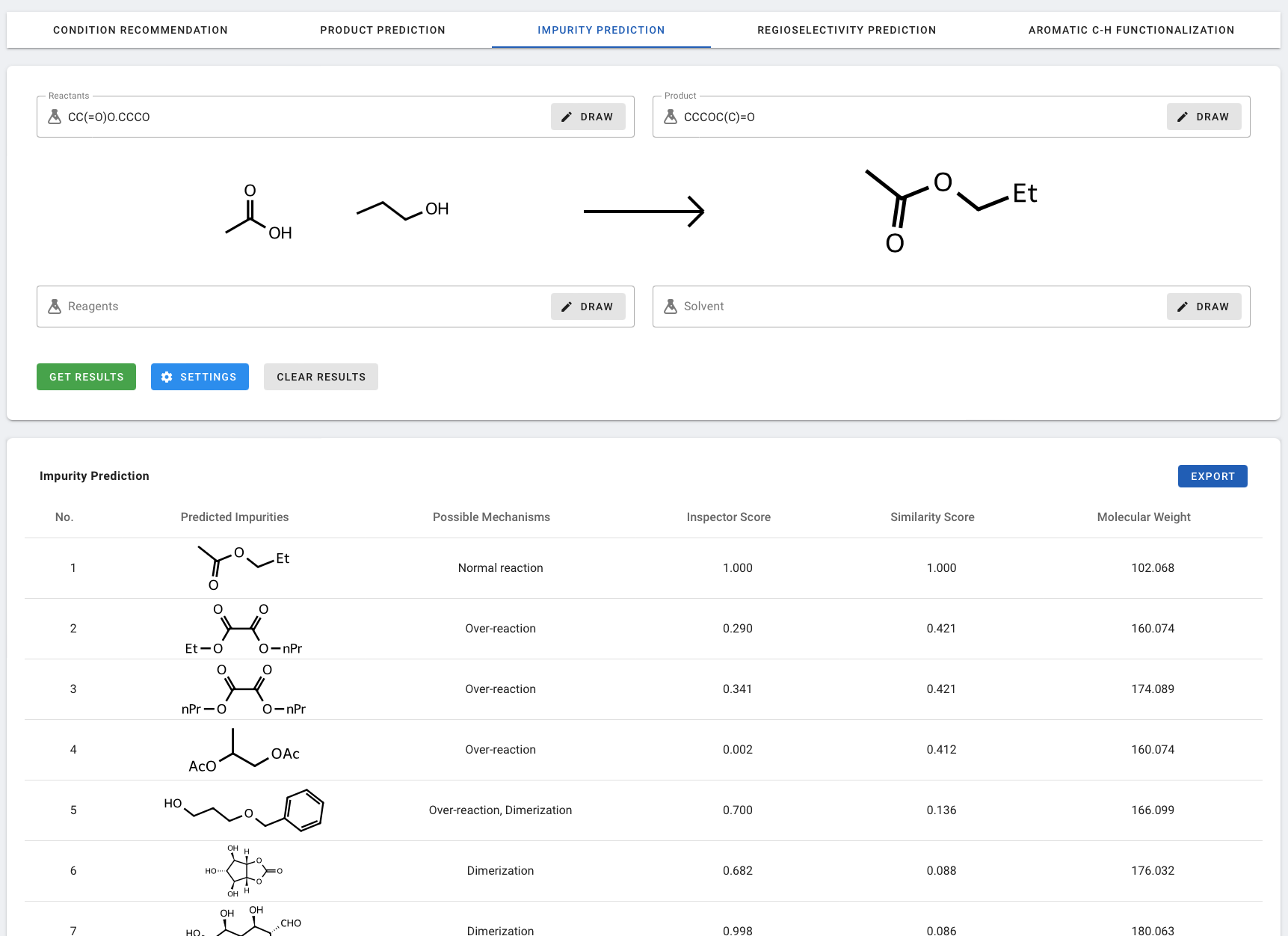
- No.: The predicted ranking provided by the model.
- Predicted Impurities: The molecular structure of the predicted impurity.
- Possible Mechanisms: Possible impurity forming mechanism(s) from Mode 1-5, as described above.
- Inspector score: Probability score from the forward prediction model.
- Similarity score: Molecular similarity with the (predicted) major product.
- Molecular Weight: The molecular weight of the predicted impurity.
How to properly use inspector score in impurity prediction:
Different impurities can be formed from different reactant & reagent sets (e.g. Minor product D is formed from ‘A+B → D’ reaction and over-reaction product E is formed from ‘A+D → E’). Thus, comparing probability scores of different impurities is not an apple-to-apple comparison when reactant & reagent sets are different and probability score cannot be used for comparing the formation likelihood of impurities.
On the other hand, comparing Fast Filter scores can give you an idea of which impurities are (roughly) more likely to be formed, but it also needs to be used with care as the following example demonstrates: Comparing Fast Filter scores of ‘A+B → D’ (Mode 1) and dimerization reaction ‘A+A → F’ (Mode 3) will be a more meaningful comparison than comparing Fast Filter scores of ‘A+B → D’ (Mode 1) and dimerization reaction ‘A+D → E’ (Mode 2). Fast Filter score of ‘A+D → E’ does not reflect the formation likelihood of D (Mode 1 product) which is crucial in overall formation likelihood of over-reaction product E.
Inspector score is used with inspection threshold to filter out unlikely reactions during impurity prediction. In general, using probability score yields better filtering results. If you are focusing on creating a more complete set of potential impurities, use probability score. If you want to roughly compare the formation likelihood of different impurities, use Fast Filter score (with care).
Regioselectivity Prediction
Regioselectivity Prediction predicts the selectivity of regio-selective reactions. The QM-GNN model combines WLN graph encoding with predicted quantum descriptors as input to a multitask neural network.
- Enter O=CC1CCCCC1.c1ccc(C2CCCCC2)nc1 as the Reactants and c1cc(C2CCCCC2)nc(C2CCCCC2)c1 as the Products.
- Click Settings.
- Do not map reagents: When turned off, reagents will be mapped and considered as a potential participant in the reaction. Default is off.
- Click Get results to start prediction.
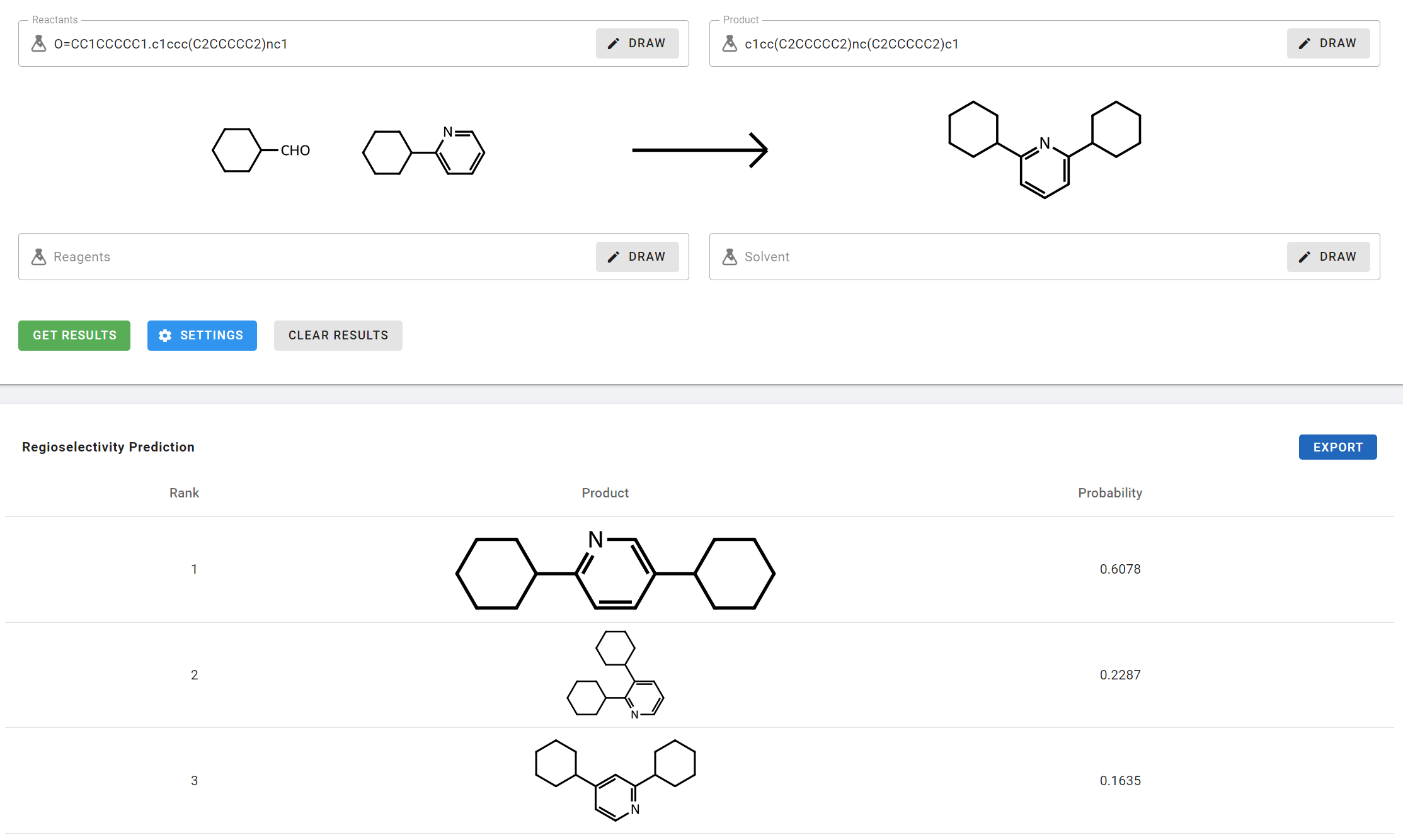
The results table lists the predicted regioisomer products based on probability:
- Rank: The predicted ranking provided by the model
- Product: The molecular structure of product
- Probability: Probability score from the forward prediction model.
Results can be exported to a csv file by clicking EXPORT button.
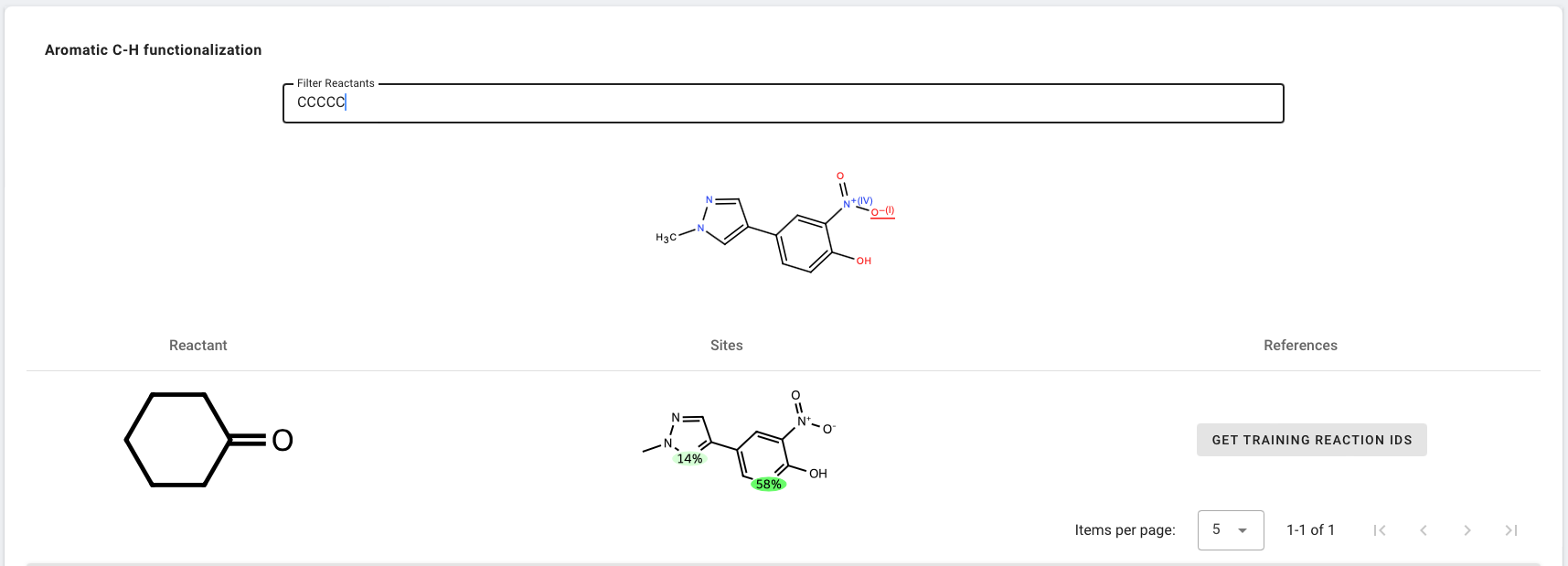
Aromatic C-H Functionalization
Aromatic C-H Functionalization predicts site selectivity of aromatic C-H functionalization reactions with a multitask neural network that uses WLN graph encoding.
- Enter
Cn1cc(-c2ccc(O)c([N+](=O)[O-])c2)cn1as the Reactants. - Click Get results to start prediction.
In the results table, you can view the probability of C-H functionalization with a specific reactant. 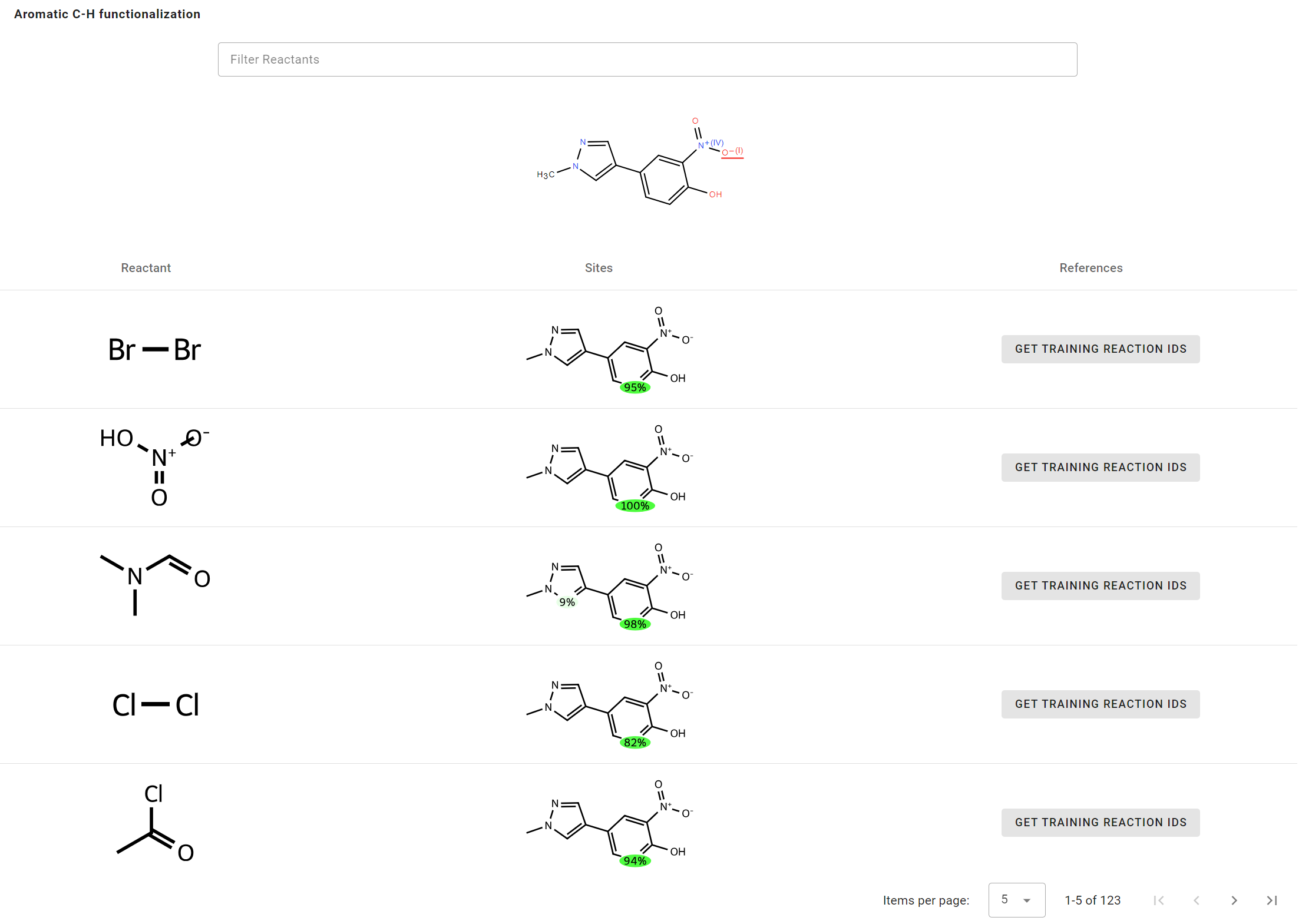
If you click any aromatic atom (highlighted in blue) in molecule just above the table (highlighted in red), you can see the most probable reactant(s) that can functionalize that site. 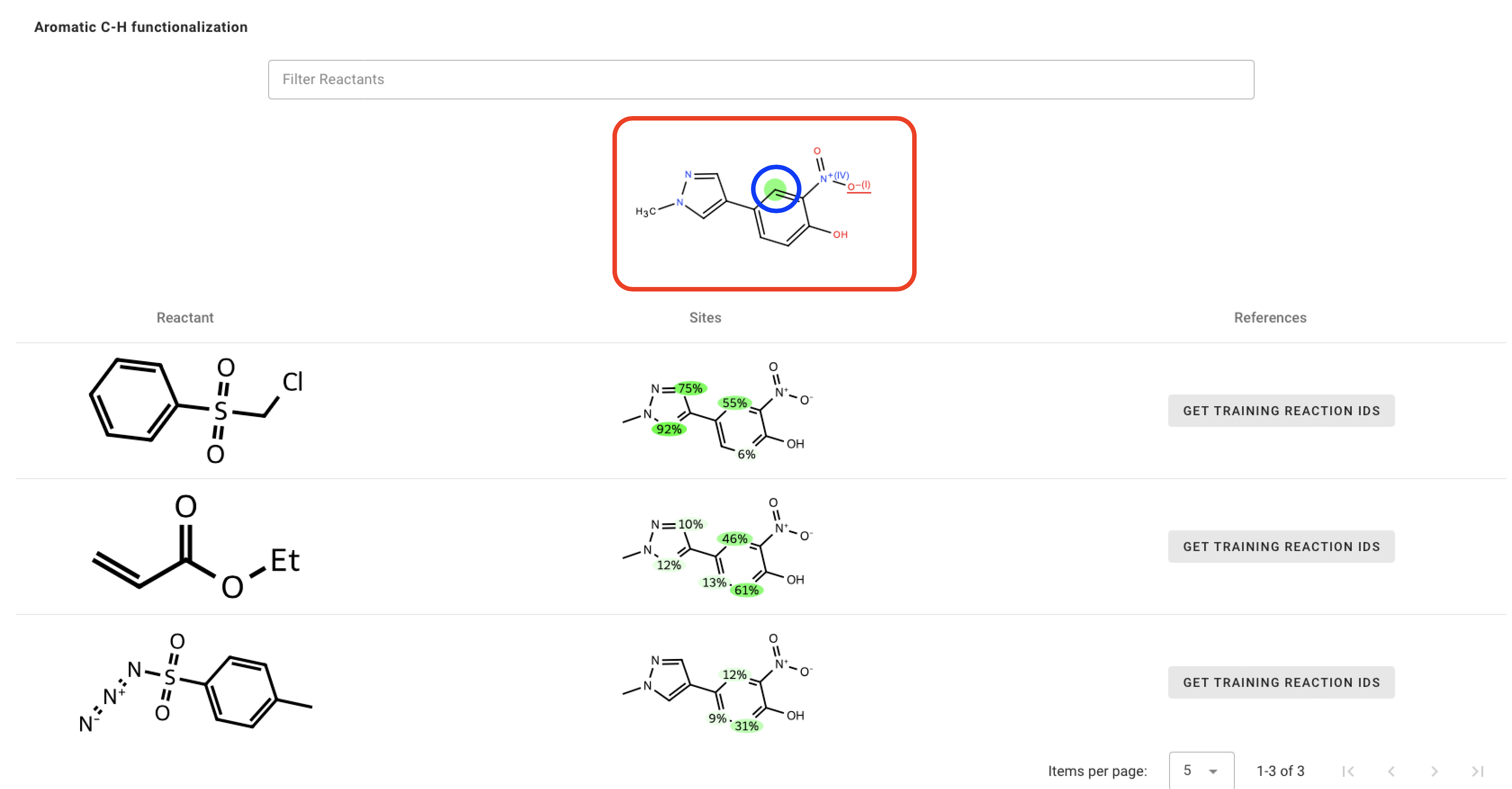
If you type any reactants SMILES in the Filter Reactants box, you can see the site-selectivity score with that specific reactant (CCCCCC in the example below).